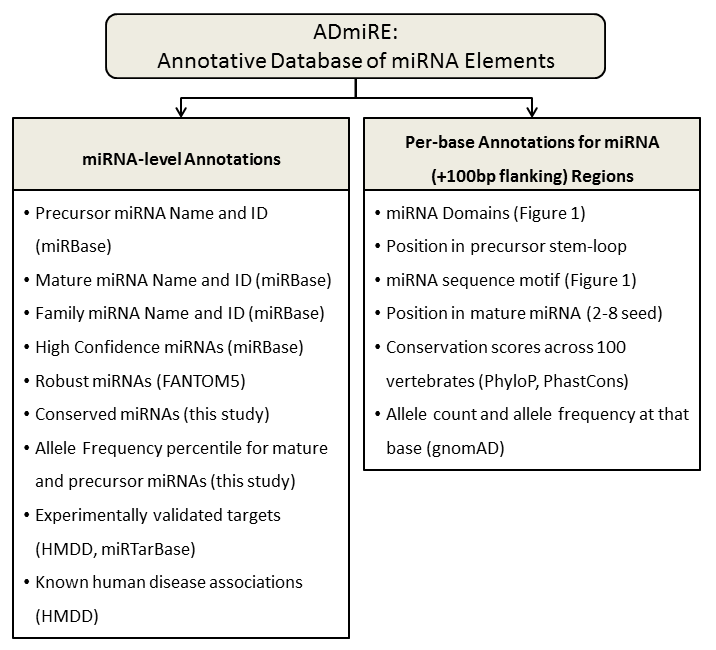
| 1 |
MIRNA |
Precursor Name (miRBase) |
| 2 |
PRE_ID |
Precursor ID (miRBase) |
| 3 |
Family_Name |
miRNA Family Name (miRBase) |
| 4 |
Family_ID |
miRNA Family ID (miRBase) |
| 5 |
Mature_Name |
Mature Name (miRBase) (if applicable) |
| 6 |
Mature_ID |
Mature ID (miRBase) (if applicable) |
| 7 |
High_Confidence |
High Confidence (miRBase) |
| 8 |
Conserved_ADmiRE |
Conserved ADmiRE (This study) |
| 9 |
Robust_FANTOM5 |
Robust miRNA (FANTOM5 project) |
| 10 |
HMDD_Targets.PMID |
Curated experiementally validated targets (PubMed ID) (HMDD) |
| 11 |
HMDD_Disease.PMID |
Known associated disease (HMDD) |
| 12 |
miRTarBase_Validated.Targets |
Curated experimentally validated targets (miRTarBase) |
| 13 |
miRTarBase_TargetGene.Validation.type |
Method of target validation (miRTarBase) |
| 14 |
miRTarBase_Reference.PMID |
Publication of target validation (miRTarBase) |
| 15 |
gnomad_af_precursor_quantile |
AF percentile from this study with mean AF across precursor miRNAs |
| 16 |
gnomad_af_mature_quantile |
AF percentile from this study with mean AF across mature miRNAs |
| 17 |
gnomad_af_family_quantile |
AF percentile from this study with mean AF across all mature miRNAs in a family |
| 18 |
MIRNA_Feature |
miRNA Domain (miRBase- Figure 1) |
| 19 |
Precursor_Pos |
Base position in precursor domain |
| 20 |
Pred_Motif |
Predicted Motif in Precursor (CNNC, basalUG, UGU, UGUG, loop) |
| 21 |
Mature_Pos |
Base position in mature domain |
| 22 |
PhyloP_100way |
PhyloP 100way- Vertebrate scores |
| 23 |
PhastCons_100way |
Phastcons 100way- Vertebrate scores |
| 24 |
gnomAD_Count |
gnomAD AC for all alleles at the position |
| 25 |
gnomAD_MAF |
gnomAD AF for all alleles at the position |